Linear Interpolation#
The simplest interpolation we can imagine is doing connect-the-dots, and simply connecting our data samples with straight lines.
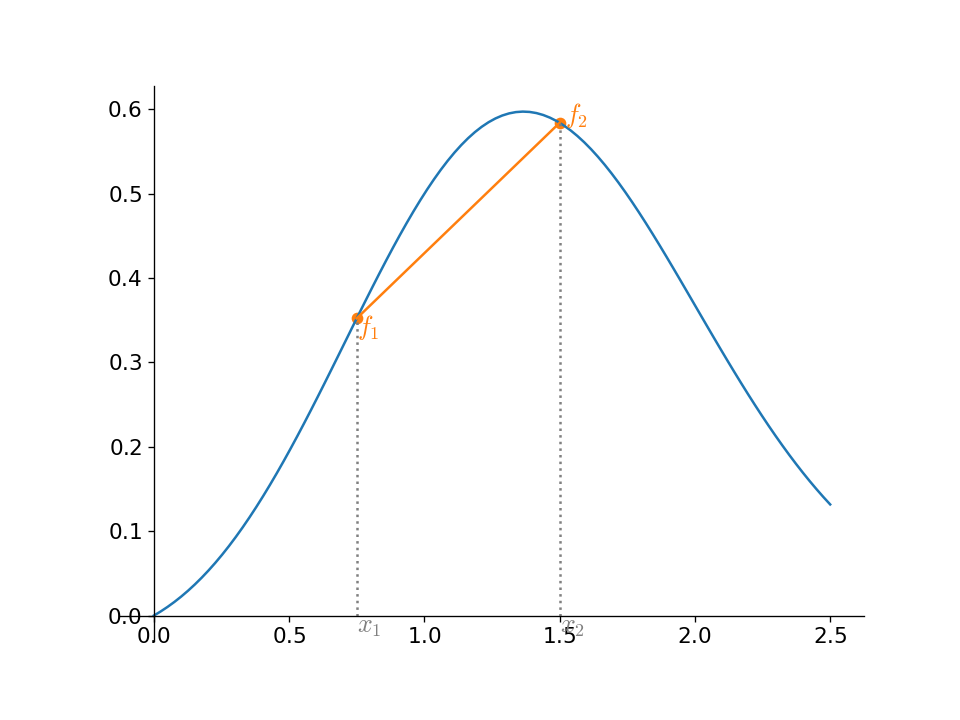
As we see, this only gets the correct values at the end points (for a general function), but it has the nice property that it does not introduce any new minima or maxima.
Given some N points sampling a function, \((x_i, f_i)\), the interpolant connecting the points \(i\) and \(i+1\) is:
for \(x_i \le x \le x_{i+1}\)
For N points, we have \(N-1\) interpolants.
Piecewise interpolation#
This is an example of piecewise interpolation. We don’t try to fit all of the points with a single polynomial, but instead fit a subset of points with a low-order polynomial (in this case, a line).
Example
Let’s write a function to tabulate \(f(x) = x \sin(x)\) on \([0, 5]\) and then use linear interpolation to get the function values.
How many function samples do we need to get a 1% error?
import numpy as np
import matplotlib.pyplot as plt
def f(x):
return x * np.sin(x)
def sample_function(func, npts, *, xmin=0.0, xmax=10.0):
xx = np.linspace(xmin, xmax, npts)
return xx, func(xx)
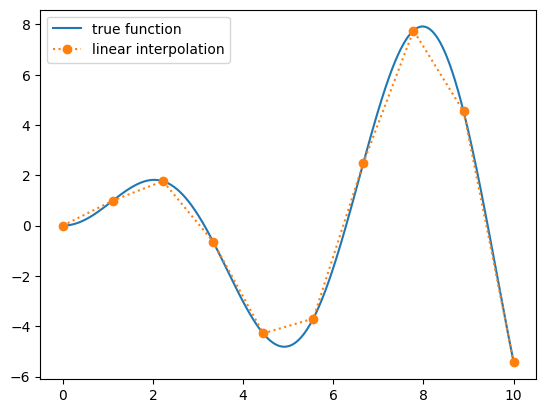
Let’s look at this function and 10 samples
fig, ax = plt.subplots()
xmin = 0.0
xmax = 10.0
N = 10
x_fine = np.linspace(xmin, xmax, 1000)
x_sample, f_sample = sample_function(f, N, xmin=xmin, xmax=xmax)
ax.plot(x_fine, f(x_fine), label="true function")
ax.plot(x_sample, f_sample, marker="o", ls=":", label="linear interpolation")
ax.legend()
<matplotlib.legend.Legend at 0x7f48b842e120>
Now let’s define our interpolate function. This will take the location and values where the function is sampled, (xv, fv), and the location where we want to approximate the value of the underlying function, x0.
def interpolate(x0, xv, fv):
# find first x[i] > x0
idx = np.argwhere(xv > x0)[0][0]
# we want to use this point and the one to the left
# we'll shift idx to point to the left point
idx = max(0, idx-1)
slope = (fv[idx+1] - fv[idx]) / (xv[idx+1] - xv[idx])
return slope * (x0 - xv[idx]) + fv[idx]
Let’s just look at \(x_0 = 4.5\) (this is near a minimum) and try different number of points and measure the error
x0 = 4.5
for n in [5, 10, 20, 40, 80, 160]:
xv, fv = sample_function(f, n)
finterp = interpolate(x0, xv, fv)
fexact = f(x0)
err = np.abs(finterp - fexact)
print(f"{n:4} points, error = {err}")
5 points, error = 0.8624245028923605
10 points, error = 0.14257394829953363
20 points, error = 0.1333081368280027
40 points, error = 0.03204962648664189
80 points, error = 0.007851522573614211
160 points, error = 0.0019427475977087383
In this case, after 80 points, our error is < 1%.
We also see that we appear to converge slightly better than 2nd order
C++ implementation#
Here’s a C++ version that follows the same ideas:
#include <iostream>
#include <cmath>
#include <vector>
#include <functional>
#include <print>
// our test function
double f(double x) {
return x * std::sin(x);
}
// a function that will produce N samples of the input func(x)
std::pair<std::vector<double>, std::vector<double>>
sample_function(std::function<double(double)> func,
int npts, double xmin, double xmax) {
std::vector<double> xv(npts, 0.0);
std::vector<double> fv(npts, 0.0);
double dx = (xmax - xmin) / static_cast<double>(npts - 1);
for (int i = 0; i < npts; ++i) {
xv[i] = xmin + static_cast<double>(i) * dx;
fv[i] = func(xv[i]);
}
return {xv, fv};
}
// the main linear intepolation function
double interpolate(double x0,
const std::vector<double>& xv,
const std::vector<double>& fv) {
// find the index i where xv[i] > x0
int idx = -1;
for (std::size_t i = 0; i < xv.size()-1; ++i) {
if (x0 > xv[i] && x0 < xv[i+1]) {
idx = i;
break;
}
}
if (idx == -1) {
std::cout << "x0 is not in range of xv" << std::endl;
}
double slope = (fv[idx+1] - fv[idx]) / (xv[idx+1] - xv[idx]);
return slope * (x0 - xv[idx]) + fv[idx];
}
// test driver
int main() {
// we want to find the N such that the error in our interpolation
// is < 1%. We'll just consider a bunch of different sizes for N
double xmin{0.0};
double xmax{10.0};
double x0{4.5};
for (auto N : {5, 10, 20, 40, 80, 160}) {
const auto [xv, fv] = sample_function(f, N, xmin, xmax);
auto finterp = interpolate(x0, xv, fv);
auto fexact = f(x0);
double err = std::abs(finterp - fexact);
std::println("{:4} points, error = {:7.5f}", N, err);
}
}


