Implementing Relaxation#
import numpy as np
import matplotlib.pyplot as plt
Let’s start by writing a grid class that holds the solution and RHS and knows how to fill ghost cells. Much of this class is identical to what we’ve done before.
class Grid:
def __init__(self, nx, ng=1, xmin=0, xmax=1,
bc_left_type="dirichlet", bc_left_val=0.0,
bc_right_type="dirichlet", bc_right_val=0.0):
self.xmin = xmin
self.xmax = xmax
self.ng = ng
self.nx = nx
self.bc_left_type = bc_left_type
self.bc_left_val = bc_left_val
self.bc_right_type = bc_right_type
self.bc_right_val = bc_right_val
# python is zero-based. Make easy integers to know where the
# real data lives
self.ilo = ng
self.ihi = ng+nx-1
# physical coords -- cell-centered
self.dx = (xmax - xmin)/(nx)
self.x = xmin + (np.arange(nx+2*ng)-ng+0.5)*self.dx
# storage for the solution
self.phi = self.scratch_array()
self.f = self.scratch_array()
def scratch_array(self):
"""return a scratch array dimensioned for our grid """
return np.zeros((self.nx+2*self.ng), dtype=np.float64)
def fill_bcs(self):
"""fill the boundary conditions on phi"""
# we only deal with a single ghost cell here
# left
if self.bc_left_type.lower() == "dirichlet":
self.phi[self.ilo-1] = 2 * self.bc_left_val - self.phi[self.ilo]
elif self.bc_left_type.lower() == "neumann":
self.phi[self.ilo-1] = self.phi[self.ilo] - self.dx * self.bc_left_val
else:
raise ValueError("invalid bc_left_type")
# right
if self.bc_right_type.lower() == "dirichlet":
self.phi[self.ihi+1] = 2 * self.bc_right_val - self.phi[self.ihi]
elif self.bc_right_type.lower() == "neumann":
self.phi[self.ihi+1] = self.phi[self.ihi] - self.dx * self.bc_right_val
else:
raise ValueError("invalid bc_right_type")
Next we’ll write a smoother routine, that does a single pass of red-black G-S. Notice that we first update zones ilo, ilo+2, ilo+4, … and then we need to fill the ghost cells with our boundary conditions again. Then we do the next sweep, this time updating ilo+1, ilo+3, ilo+5, …
def smooth(g):
"""perform red-black Gauss-Seidel smoothing"""
g.fill_bcs()
g.phi[g.ilo:g.ihi+1:2] = 0.5 * (-g.dx * g.dx * g.f[g.ilo:g.ihi+1:2] +
g.phi[g.ilo+1:g.ihi+2:2] + g.phi[g.ilo-1:g.ihi:2])
g.fill_bcs()
g.phi[g.ilo+1:g.ihi+1:2] = 0.5 * (-g.dx * g.dx * g.f[g.ilo+1:g.ihi+1:2] +
g.phi[g.ilo+2:g.ihi+2:2] + g.phi[g.ilo:g.ihi:2])
Test problem#
Now we are ready to try this out. Let’s do the problem:
on \([0, 1]\) with homogeneous Dirichlet BCs.
The solution is:
Let’s create functions that initialize the RHS and the analytic solution.
def analytic(x):
return -np.sin(x) + x * np.sin(1.0)
def f(x):
return np.sin(x)
Now let’s create the grid—we’ll use 128 points to start. The defaults for the grid class already set homogeneous Dirichlet BCs.
g = Grid(128)
Now we need to initialize \(\phi\) and \(f\).
For \(\phi\), we may as well just initialize it to \(0\) (which is already done for us by the grid class), since usually we don’t know what the answer is.
g.f[:] = f(g.x)
Relaxing#
Now we can do relaxation / smoothing. We don’t know how many smoothing iterations we need, and we also haven’t yet discussed how to determine what the error is / when to stop. So we’ll look at the result after a fixed number of smoothing iterations.
fig, ax = plt.subplots()
niters = [1, 10, 100, 1000, 10000]
for n in niters:
# smooth n times
for _ in range(n):
smooth(g)
ax.plot(g.x[g.ilo:g.ihi+1], g.phi[g.ilo:g.ihi+1], label=f"N = {n}")
# reset phi to 0 for the next number of iterations
g.phi[:] = 0.0
ax.plot(g.x[g.ilo:g.ihi+1], analytic(g.x[g.ilo:g.ihi+1]), ls=":",
color="0.5", label="analytic")
ax.legend()
<matplotlib.legend.Legend at 0x7f6442b2e270>
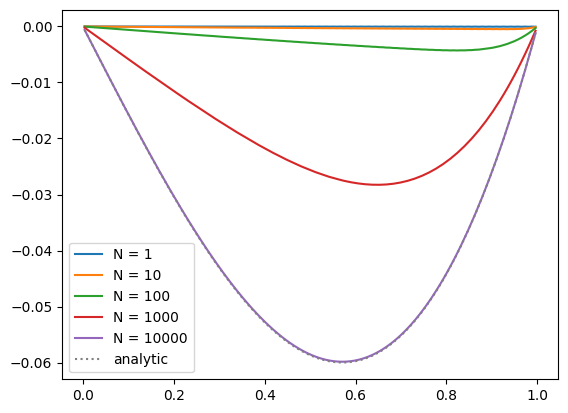
We see that as we increase the number of smoothing iterations, we get closer to the analytic solution. Also note that the solution appears to be respecting our boundary conditions well.
It looks like only after 10000 iterations do we get close to the solution.
Important
We haven’t yet figured out how to tell how much relaxing is enough…


