Fitting to a Line#
Let’s consider the case of fitting data to a line. Our model has the form:
and our fit appears as:
We want to minimize \(\chi^2(a_1, a_2)\).
We will actually care about the reduced chi-square, which is scaled by the number of degrees of freedom, \(N - M\), where \(N\) is the number of data points and \(M\) is the number of fitting parameters.
Tip
Generally we want the \(\chi^2 < 1\) for a fit to be considered “good”.
We start by differentiating with respect to the fit parameters and setting the derivatives to zero:
Separating the terms, we have:
This is a linear system with 2 equations and 2 unknowns.
Let’s define:
Then our system is:
We can solve this easily:
Example data#
Let’s make some sample data that we perturb with Gaussian-normalized noise. We’ll use the NumPy standard_normal function.
Tip
In C++, you can generate random numbers in a normal distribution via std::normal_distribution.
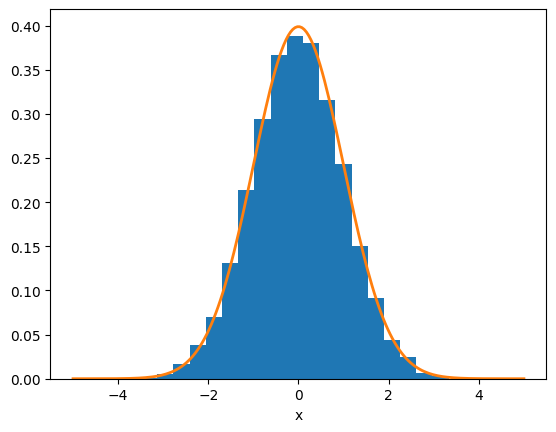
Let’s first see how this works. Let’s take a large number of samples and also plot a Gaussian distribution:
import numpy as np
import matplotlib.pyplot as plt
N = 10000
rng = np.random.default_rng()
r = rng.standard_normal(N)
fig, ax = plt.subplots()
ax.hist(r, density=True, bins=20)
x = np.linspace(-5, 5, 200)
sigma = 1.0
ax.plot(x, np.exp(-x**2/(2*sigma**2)) / (sigma*np.sqrt(2.0*np.pi)), lw=2)
ax.set_xlabel("x")
Text(0.5, 0, 'x')
Now we can make some experimental data. This will be data that follows a line, but is perturbed by a Gaussian-normalized random number, to give it some experimental error.
def y_experiment(a1, a2, sigma, x):
""" return the experimental data and error in a linear + random
fashion; a1 is the intercept, a2 is the slope, and sigma is the
error scale"""
N = len(x)
rng = np.random.default_rng()
r = sigma * rng.standard_normal(N)
yerr = sigma * np.ones_like(rng)
return a1 + a2*x + r, yerr
Now we can make the data that we want to fit to.
# number of data points
N = 40
# one-sigma error
sigma = 25.0
# independent variable
x = np.linspace(0.0, 100, N)
y, yerr = y_experiment(10.0, 3.0, sigma, x)
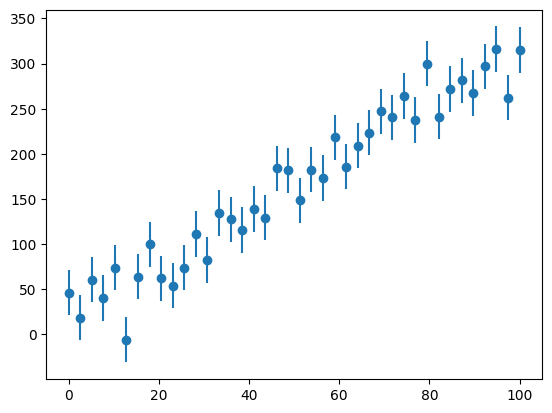
Let’s look at our “experiment”:
fig, ax = plt.subplots()
ax.errorbar(x, y, yerr=yerr, fmt="o")
<ErrorbarContainer object of 3 artists>
Now we can write a function to do the fitting
def linear_regression(x, y, yerr):
"""fit data (x_i, y_i) with errors {yerr_i} to a line"""
N = len(x)
C = np.sum(1.0 / yerr**2)
S_x = np.sum(x / yerr**2)
S_x2 = np.sum(x * x / yerr**2)
S_y = np.sum(y / yerr**2)
S_xy = np.sum(x * y / yerr**2)
a2 = (C * S_xy - S_x * S_y)/(C * S_x2 - S_x**2)
a1 = (S_y * S_x2 - S_xy * S_x) / (C * S_x2 - S_x**2)
chisq = np.sum((a1 + a2 * x - y)**2 / yerr**2)
chisq /= N-2
return a1, a2, chisq
a1, a2, chisq = linear_regression(x, y, yerr)
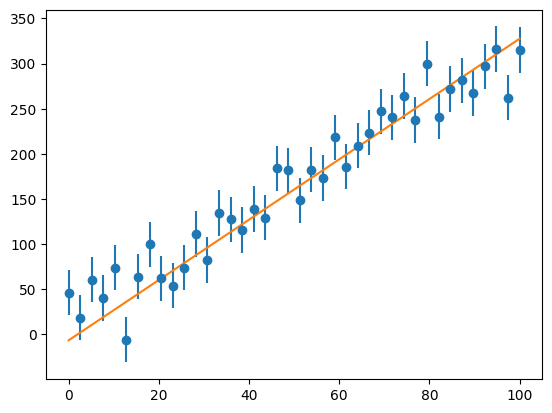
Now we can look at how well our fit does:
ax.plot(x, a1 + a2*x)
fig
Pathologies#
We need to be careful to not over-interpret a fit. Consider the Anscombe’s quartet. These 4 very different datasets all have the same linear fit (to a few digits significance).


