Nonlinear Fitting#
What about the case of fitting to a function where the fit parameters enter in a nonlinear fashion? For example:
Note
One trick that is often used for something like this is to transform the data. So instead of fitting the data \((x_i, y_i)\), you instead fit \((x_i, \log y_i)\), and then our fitting function is:
which is linear.
However, when there are errors associated with the \(y_i\), the errors do not necessarily transform the correct way when you take the logarithm.
Let’s look at how we would fit directly to a nonlinear function.
We’ll minimize the same fitting function:
with fitting parameters \({\bf a} = (a_1, \ldots, a_M)^\intercal\).
Now we take the derivatives with respect to each parameter, \(a_k\):
Let’s define \(g_k \equiv {\partial \chi^2}/{\partial a_k}\), then we have
This is a nonlinear system of \(M\) equations and \(M\) unknowns. We can solve this using the same multivariate Newton’s method we looked at before:
Take an initial guess at the fit parameters, \({\bf a}^{(k)}\)
Solve the system \({\bf J}\delta {\bf a} = -{\bf g}\), where \(J_{ij} = \partial g_i/\partial a_j\) is the Jacobian
Correct the initial guess, \({\bf a}^{(k+1)} = {\bf a}^{(k)} + \delta {\bf a}\)
Warning
As we’ve seen with Newton’s method, convergence will be very sensitive to the initial guess.
import numpy as np
import matplotlib.pyplot as plt
Fitting an exponential#
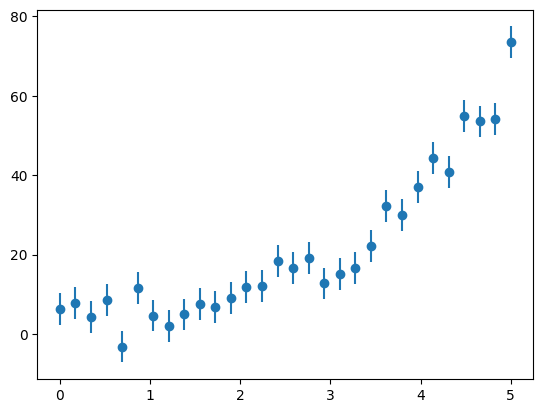
Let’s try this out on data that is constructed to follow an exponential trend.
First let’s construct the data, and perturb it with some errors. We’ll take the form:
# make up some experimental data
a0 = 2.5
a1 = 2./3.
sigma = 4.0
N = 30
x = np.linspace(0.0, 5.0, N)
We will do a Gaussian-normal sampling of the error (with width \(\sigma\)), but in the fit, the uncertainty is just \(\sigma\).
rng = np.random.default_rng()
r = sigma * rng.standard_normal(N)
y = a0 * np.exp(a1 * x) + r
yerr = sigma * np.ones_like(r)
fig, ax = plt.subplots()
ax.errorbar(x, y, yerr=yerr, fmt="o")
<ErrorbarContainer object of 3 artists>
Now, let’s compute our vector \({\bf g}\) that we will zero:
We can divide out the \(-2\) in each expression. We’ll keep the overall \(a_0\) in the expression, to deal with the case where it might be \(0\). This gives:
Let’s write a function to compute this:
def g(x, y, yerr, a):
"""compute the nonlinear functions we minimize. Here a is the vector
of fit parameters"""
a0, a1 = a
g0 = np.sum(np.exp(a1 * x) * (y - a0 * np.exp(a1 * x)) / yerr**2)
g1 = a0 * np.sum(x * np.exp(a1 * x) * (y - a0 * np.exp(a1 * x)) / yerr**2)
return np.array([g0, g1])
We also need the Jacobian. We could either compute this numerically, via differencing, or analytically. We’ll do the latter.
Notice that the Jacobian is symmetric:
This is called the Hessian matrix.
Let’s write this function:
def jac(x, y, yerr, a):
""" compute the Jacobian of the function g"""
a0, a1 = a
dg0da0 = -np.sum(np.exp(2.0 * a1 * x) / yerr**2)
dg0da1 = np.sum(x * np.exp(a1 * x) * (y - 2.0 * a0 * np.exp(a1 * x)) / yerr**2)
dg1da0 = dg0da1
dg1da1 = np.sum(a0 * x**2 * np.exp(a1 * x) * (y - 2.0 * a0 * np.exp(a1 * x)) / yerr**2)
return np.array([[dg0da0, dg0da1],
[dg1da0, dg1da1]])
Tip
Convergence here is very sensitive to the initial guess. We’ll do two modifications to deal with this:
we add a cap on the number of iterations allowed prevent this from being an infinite loop if we can’t converge
we prevent our \([a_0, a_1]\) from changing by more than 20% per iteration
def fit(aguess, x, y, yerr, tol=1.e-5):
""" aguess is the initial guess to our fit parameters. x and y
are the vector of points that we are fitting to, and yerr are
the errors in y"""
avec = aguess.copy()
max_steps = 100
err = 1.e100
n = 0
while err > tol and n < max_steps:
# get the jacobian
J = jac(x, y, yerr, avec)
# get the current function values
gv = g(x, y, yerr, avec)
# solve for the correction: J da = -g
da = np.linalg.solve(J, -gv)
# limit the change
avec = np.clip(avec + da, 0.8 * avec, 1.2 * avec)
err = np.max(np.abs(da/avec))
n += 1
return avec
# initial guesses
aguess = np.array([2.0, 1.0])
# fit
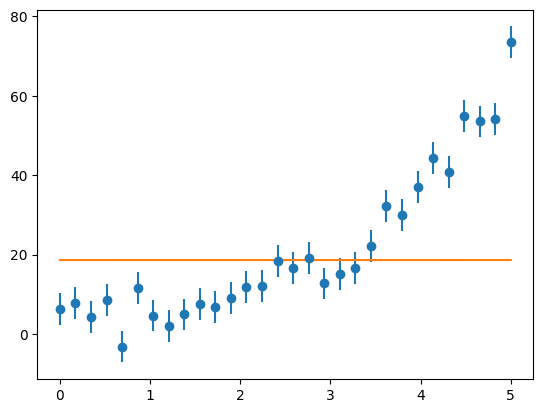
afit = fit(aguess, x, y, yerr)
afit
array([1.85996851e+01, 3.97613147e-05])
ax.plot(x, afit[0] * np.exp(afit[1] *x))
fig
Is it a minimum?#
We just found an extrema. Let’s plot the surface around our fit parameters to see if it looks like a minimum
npts = 100
a0v = np.linspace(0.5 * afit[0], 2.0 * afit[0], npts)
a1v = np.linspace(0.5 * afit[1], 2.0 * afit[1], npts)
def chisq(a0, a1, x, y, yerr):
return np.sum((y - a0 * np.exp(a1 * x))**2 / yerr**2)
chisq(afit[0], afit[1], x, y, yerr)
np.float64(679.5434792130577)
c2 = np.zeros((npts, npts), dtype=np.float64)
for i, a0 in enumerate(a0v):
for j, a1 in enumerate(a1v):
c2[i, j] = chisq(a0, a1, x, y, yerr)
Now we’ll plot the (log of) the \(\chi^2\)
fig, ax = plt.subplots()
# we need to transpose to put a0 on the horizontal
# we use origin = lower to have the origin at the lower left
im = ax.imshow(np.log10(c2).T,
origin="lower",
extent=[a0v[0], a0v[-1], a1v[0], a1v[-1]])
fig.colorbar(im, ax=ax, orientation="horizontal")
ax.scatter([afit[0]], [afit[1]], color="r", marker="x")
ax.set_xlabel("$a_0$")
ax.set_ylabel("$a_1$")
Text(0, 0.5, '$a_1$')
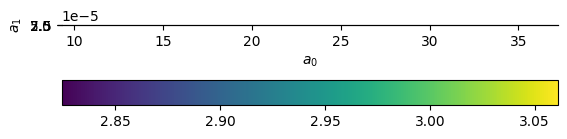
It looks like there is a very broad minimum there.
Note
Consider if we tried to add another parameter, fitting to:
here \(a_2\) enters the same way as \(a_0\), which would give a singular matrix, and make our solution unstable.


