Homework 6 solutions#
import numpy as np
import matplotlib.pyplot as plt
1. Cepheids#
We want to fit a line to Cepheid period-magnitudes and find the error in the fit.
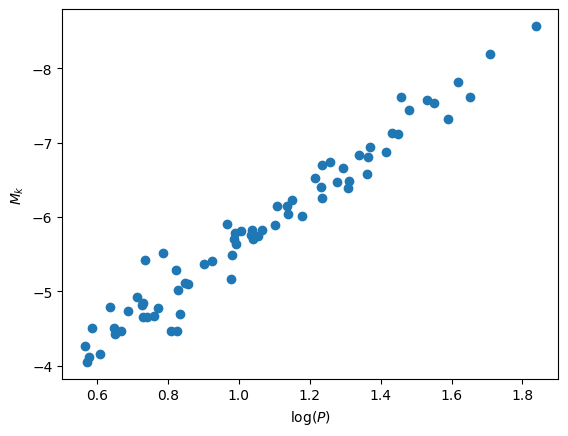
Let’s start by reading in the data and making a plot. We only care about 3 columns, “logP”, “MK”, and “σmM”
data = np.genfromtxt("cepheids.txt", names=True)
fig, ax = plt.subplots()
ax.scatter(data["logP"], data["MK"])
ax.set_xlabel(r"$\log(P)$")
ax.set_ylabel("$M_k$")
ax.invert_yaxis()
Storm et al. 2011 fit this data to
so our model function will be:
In class, we saw that
logP = data["logP"]
Mk = data["MK"]
err = data["σmM"]
We’ll use the general linear least squares from class:
def basis(x, M=3):
""" the basis function for the fit, x**n"""
j = np.arange(M)
return x**j
This is modified from class to return the matrix \({\bf A}^\intercal{\bf A}\).
def general_regression(x, y, yerr, M):
""" here, M is the number of fitting parameters. We will fit to
a function that is linear in the a's, using the basis functions
x**j """
N = len(x)
# construct the design matrix -- A_{ij} = Y_j(x_i)/sigma_i -- this is
# N x M. Each row corresponds to a single data point, x_i, y_i
A = np.zeros((N, M), dtype=np.float64)
for i in range(N):
A[i,:] = basis(x[i], M) / yerr[i]
# construct the MxM matrix for the linear system, A^T A:
ATA = np.transpose(A) @ A
print("condition number of A^T A:", np.linalg.cond(ATA))
# construct the RHS
b = np.transpose(A) @ (y / yerr)
# solve the system
a = np.linalg.solve(ATA, b)
# return the chisq
chisq = 0
for i in range(N):
chisq += (np.sum(a*basis(x[i], M)) - y[i])**2 / yerr[i]**2
chisq /= N-M
return a, ATA, chisq
a, ATA, chisq = general_regression(logP-1.0, Mk, err, 2)
condition number of A^T A: 8.494389613454842
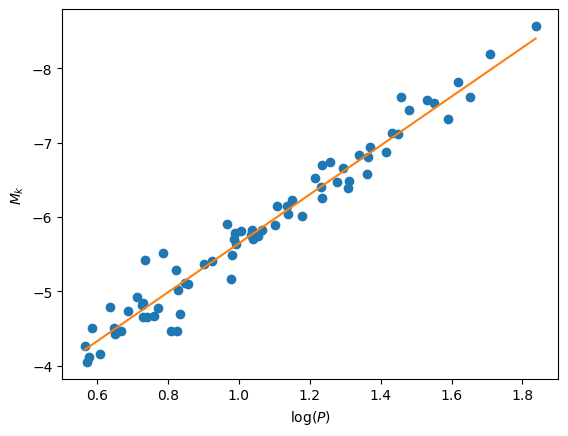
Here’s our fit:
a
array([-5.64557103, -3.28969021])
and our reduced \(\chi^2\):
This agrees well with what Storm et al. found.
chisq
np.float64(32.99816166705109)
Now let’s look at the errors. We want to compute:
ATA
array([[64368.21617536, 6779.44744347],
[ 6779.44744347, 8483.83758629]])
C = ATA**-1
C
array([[1.55356177e-05, 1.47504647e-04],
[1.47504647e-04, 1.17871186e-04]])
and then the uncertainties are the square root of the diagonals:
sigmas = np.sqrt(np.linalg.diagonal(np.linalg.inv(ATA)))
sigmas
array([0.00411865, 0.01134475])
This says that our fit parameters are:
print(f"intercept: {a[0]:6.3f} +/- {sigmas[0]:6.3f}")
print(f"slope : {a[1]:6.3f} +/- {sigmas[1]:6.3f}")
intercept: -5.646 +/- 0.004
slope : -3.290 +/- 0.011
Finally let’s plot the fit:
ax.plot(logP, (a[0] - a[1]) + a[1] * logP, color="C1")
fig
2. Interpolation#
The reaction rate for the 3-\(\alpha\) process is
def lam(T):
"""this is the triple alpha reaction rate / rho^2 Y^3"""
T8 = T/1e8
return 5.09e11 * T8**-3 * np.exp(-44.027/T8)
We want to tabulate the temperature portion of this,
this at 10 points, equally spaced in log between \(T = 10^8~\mathrm{K}\) and \(5\times 10^9~\mathrm{K}\).
First let’s create that tabulation:
Ts = np.logspace(np.log10(1.e8), np.log10(5.e9), 10)
logTs = np.log10(Ts)
logTs
array([8. , 8.18877444, 8.37754889, 8.56632333, 8.75509778,
8.94387222, 9.13264667, 9.32142111, 9.51019556, 9.69897 ])
loglams = np.log10(lam(Ts))
loglams
array([-7.41396537, -1.23984259, 2.55813006, 4.81759586, 6.08091109,
6.69923995, 6.89995379, 6.83027077, 6.58551152, 6.22739411])
Now, given our array of \(\log(T)\) and \(\log(\lambda)\), we want to do cubic Lagrange interpolation on the 4 points that surround the desired value, \((\log(T))_0\).
Given 4 points, \((\log(T))_i\), \((\log(T))_{i+1}\), \((\log(T))_{i+2}\), \((\log(T))_{i+3}\), we should choose \(i\) such that
e.g., there should be 2 points on each side of our desired temperature, except at the start or end of the sequence, where this is not possible, so we would just need to use the closest 4 points to the desired temperature.
Here’s a function that finds that index:
def find_index(logTs, logT0):
"""find i such that logTs[i+1] <= logT0 <= logTs[i+2]"""
return max(0, min(len(logTs)-4, np.argwhere(logTs > logT0)[0][0] - 2))
To test this, here’s a case where we are near the beginning of the tabulation:
find_index(logTs, 8.1)
0
and heres a test where we are in the middle:
find_index(logTs, 8.8)
np.int64(3)
Comparing to the logTs array above, both give the result we want.
Here’s our Lagrange interpolation class from lecture. We will use it in a slightly different fashion though. Instead of passing in the entire set of data, we will only pass in the 4 points centered on the value at which we wish to interpolate.
class LagrangePoly:
""" a general class for creating a Lagrange polynomial
representation of a function """
def __init__(self, xp, fp):
self.xp = xp
self.fp = fp
def evalf(self, x0):
""" given a point x0 and a function func, fit a Lagrange
polynomial through the control points and return the
interpolated value at x """
f = 0.0
# sum over points
for m in range(len(self.xp)):
# create the Lagrange basis polynomial for point m
l = 1
for n in range(len(self.xp)):
if n == m:
continue
l *= (x0 - self.xp[n])/(self.xp[m] - self.xp[n])
f += self.fp[m]*l
return f
logT0 = 9.6
idx = find_index(logTs, logT0)
l = LagrangePoly(logTs[idx:idx+4], loglams[idx:idx+4])
10**l.evalf(logT0)
np.float64(2663771.7432654747)
Now we can test this out by randomly trying points to interpolate
rng = np.random.default_rng()
max_err = -1
for i in range(25):
logT0 = rng.uniform(np.log10(1.e8), np.log10(5.e9))
true_rate = lam(10**logT0)
idx = find_index(logTs, logT0)
l = LagrangePoly(logTs[idx:idx+4], loglams[idx:idx+4])
interp_rate = 10.0**l.evalf(logT0)
print(f"T = {10.0**logT0:9.4g}; rate: true = {true_rate:10.5g}, interp = {interp_rate:10.5g}")
max_err = max(max_err, abs(interp_rate - true_rate)/true_rate)
print(f"maximum relative error = {max_err:9.4g}")
T = 1.928e+08; rate: true = 8.5731, interp = 8.7417
T = 1.514e+09; rate: true = 8.0059e+06, interp = 8.0192e+06
T = 2.869e+09; rate: true = 4.6465e+06, interp = 4.6517e+06
T = 4.209e+09; rate: true = 2.3987e+06, interp = 2.3932e+06
T = 1.245e+08; rate: true = 0.0001166, interp = 0.00011256
T = 8.668e+08; rate: true = 4.8651e+06, interp = 4.8678e+06
T = 1.088e+08; rate: true = 1.0656e-06, interp = 1.0331e-06
T = 7.392e+08; rate: true = 3.2643e+06, interp = 3.2807e+06
T = 1.484e+09; rate: true = 8.016e+06, interp = 8.0275e+06
T = 1.34e+09; rate: true = 7.9155e+06, interp = 7.9182e+06
T = 8.133e+08; rate: true = 4.2166e+06, interp = 4.2288e+06
T = 7.493e+08; rate: true = 3.3962e+06, interp = 3.4126e+06
T = 2.486e+08; rate: true = 675.8, interp = 678.64
T = 2.323e+09; rate: true = 6.1025e+06, interp = 6.1088e+06
T = 2.348e+09; rate: true = 6.0299e+06, interp = 6.0365e+06
T = 8.273e+08; rate: true = 4.3903e+06, interp = 4.4005e+06
T = 1.418e+09; rate: true = 8.0034e+06, interp = 8.0097e+06
T = 6.391e+08; rate: true = 1.9876e+06, interp = 1.9958e+06
T = 3.239e+08; rate: true = 18724, interp = 18915
T = 3.416e+08; rate: true = 32237, interp = 32455
T = 3.403e+09; rate: true = 3.5416e+06, interp = 3.5395e+06
T = 1.212e+09; rate: true = 7.5625e+06, interp = 7.5816e+06
T = 3.853e+09; rate: true = 2.838e+06, interp = 2.8326e+06
T = 1.28e+08; rate: true = 0.00028087, interp = 0.00027197
T = 2.488e+08; rate: true = 681.58, interp = 684.48
maximum relative error = 0.03471


