Lagrange Interpolation#
Imagine that we have a dataset with \(N\) points, \(\{x_0, x_1, \ldots, x_{N-1}\}\) and associated function values \(\{f_0, f_1, \ldots, f_{N-1}\}\). We can write a general polynomial interpolation routine that passes through all \(N\) points using the Lagrange polynomial.
The basic idea is to create \(N\) basis functions of the form:
This has the property that:
We can then write our interpolant as:
Quadratic example#
Consider fitting a quadratic to three points:
Let’s imagine that they are evenly spaced, with a spacing \(\Delta x\).
The three basis functions are:
and the quadratic polynomial is:
Let’s implement this
import numpy as np
import matplotlib.pyplot as plt
def f_lagrange(x0, xv, fv):
""" xv, fv are the 3 points, x9 is where we want to evaluate """
dx = x[1] - x[0]
return (x0 - xv[1])*(x0-xv[2])*fv[0]/(2.0*dx**2) - \
(x0 - xv[0])*(x0-xv[2])*fv[1]/(dx**2) + \
(x0 - xv[0])*(x0-xv[1])*fv[2]/(2.0*dx**2)
We’ll look at several different sets of points. For each, we’ll plot the interpolated parabola through the 3 points.
fig, ax = plt.subplots()
# grid points where our data is
x = np.array([0.0, 0.5, 1.0])
f1 = np.array([1.0, -1.0, 1.0/3.0])
f2 = np.array([0.5, 0.5, 0.1])
f3 = np.array([-0.3, 1.3, 1.0])
xfine = np.linspace(0.0, 1.0, 200)
for farray in (f1, f2, f3):
ax.scatter(x, farray, marker="x", s=25)
ax.plot(xfine, f_lagrange(xfine, x, farray))
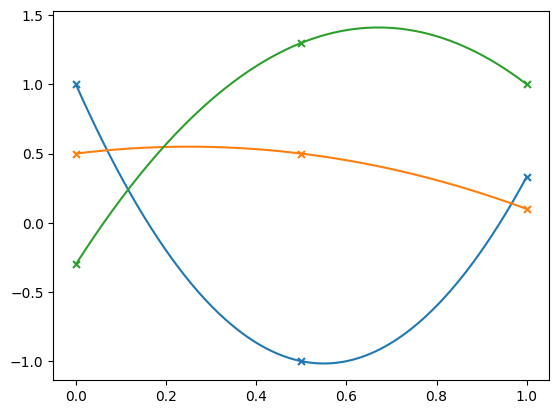
Notice that the interpolated values exceed the range of \(\{f_1, f_2, f_3\}\). For some applications we may not to introduce new extrema.
Tip
Lagrange interpolation is not very computationally efficient, since you need to recompute the basis functions for each point. There are more efficient ways to compute this, like barycentric form that are algebraically equivalent.
Runge’s phenomena#
Lagrange interpolation also suffers from Runge’s phenomenon if used with equally spaced points. This gives rise to larges osciallations at the end of the interpolating interval if we use very high-order polynomial.
Let’s write a general Lagrange interpolation class and test it out on some different functions.
class LagrangePoly:
""" a general class for creating a Lagrange polynomial
representation of a function """
def __init__(self, xmin, xmax, N, func, chebyshev=False):
self.N = N
self.xmin = xmin
self.xmax = xmax
self.func = func
if chebyshev:
k = np.arange(N) + 1
self.xp = 0.5 * (xmin + xmax) + 0.5 * (xmax - xmin) * np.cos((2 * k -1) * np.pi / (2 * N))
else:
self.xp = np.linspace(self.xmin, self.xmax, self.N)
self.fp = func(self.xp)
def evalf(self, x0):
""" given a point x0 and a function func, fit a Lagrange
polynomial through the control points and return the
interpolated value at x """
f = 0.0
# sum over points
for m in range(len(self.xp)):
# create the Lagrange basis polynomial for point m
l = 1
for n in range(len(self.xp)):
if n == m:
continue
l *= (x0 - self.xp[n])/(self.xp[m] - self.xp[n])
f += self.fp[m]*l
return f
Let’s test it out on a \(\tanh\) profile
def func(x):
return 0.5 * (1.0 + np.tanh((x - 1.0) / 0.1))
N = 11
xmin = 0.0
xmax = 2.0
l = LagrangePoly(xmin, xmax, N, func)
Let’s compute the interpolated value at a lot of points and also compute the error with respect to the exact function value.
# xx are the finely grided data that we will interpolate at to get
# the interpolated function values ff
xx = np.linspace(xmin, xmax, 200)
ff = np.zeros_like(xx)
for n, v in enumerate(xx):
ff[n] = l.evalf(v)
# exact function values at the interpolated points
fexact = func(xx)
def plot_result(xx, ff, fexact):
# compute the error
e = fexact - ff
fig = plt.figure()
ax = fig.add_subplot(211)
ax.scatter(l.xp, l.fp, marker="x", s=30)
ax.plot(xx, ff)
ax.plot(xx, fexact, color="0.5")
ax = fig.add_subplot(212)
ax.plot(xx, e)
ax.set_xlabel("x")
ax.set_ylabel("error")
fig.tight_layout()
plot_result(xx, ff, fexact)
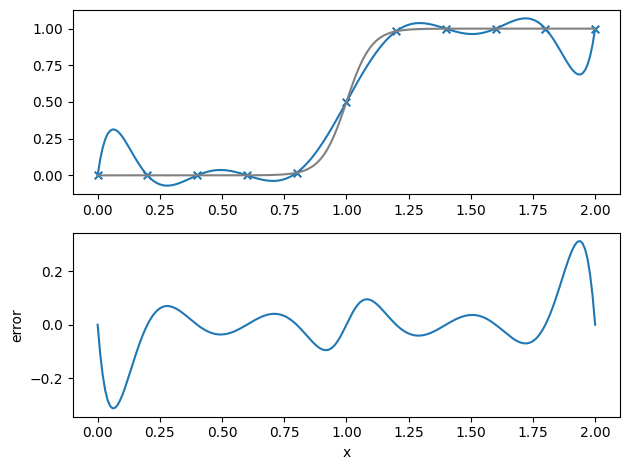
What happens as we increase the number of data samples, \(N\)?
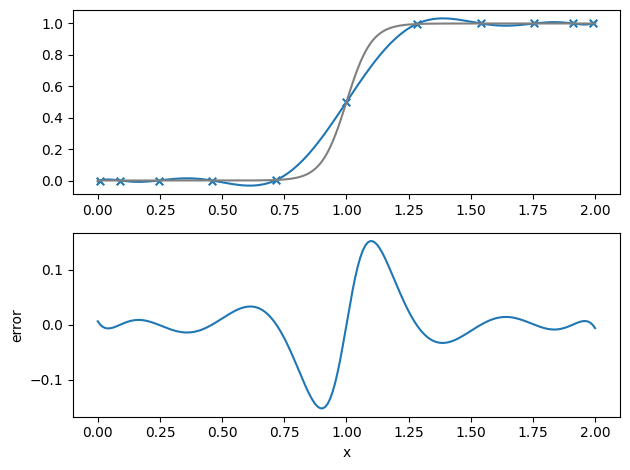
Chebyshev nodes#
If we are free to pick the points where we sample the function, we can do a lot better, and minimize the Runge’s phenomena. Sampling \(f(x)\) at the Chebyshev nodes:
where \([a, b]\) is the domain, works well.
Let’s try this out
l = LagrangePoly(xmin, xmax, N, func, chebyshev=True)
for n, v in enumerate(xx):
ff[n] = l.evalf(v)
# exact function values at the interpolated points
fexact = func(xx)
plot_result(xx, ff, fexact)
C++ implementation#
Here’s a C++ version that does Lagrange interpolation with uniform spacing in the samples:
#include <iostream>
#include <iomanip>
#include <cmath>
#include <vector>
#include <functional>
#include <fstream>
class LagrangePoly {
// create a lagrange polynomial that fits through N
// points in [xmin, xmax] sampling the function func
public:
// our data samples that we will interpolate through
std::vector<double> xp;
std::vector<double> fp;
// constructor -- this takes a function to create the
// data samples
LagrangePoly(double xmin, double xmax, int N,
std::function<double(double)> func)
: xp(N, 0.0), fp(N, 0.0)
{
double dx = (xmax - xmin) / static_cast<double>(N-1);
for (int i = 0; i < N; ++i) {
xp[i] = xmin + static_cast<double>(i) * dx;
fp[i] = func(xp[i]);
}
}
// construct and evaluate the Lagrange polynomial at x0
double evalf(double x0) {
double f{};
for (std::size_t m = 0; m < xp.size(); ++m) {
// create the Lagrange basis function for point m
double l{1};
for (std::size_t n = 0; n < xp.size(); ++n) {
if (n == m) {
continue;
}
l *= (x0 - xp[n]) / (xp[m] - xp[n]);
}
f += fp[m] * l;
}
return f;
}
};
// a test function
double f_test(double x) {
// a simple tanh profile
return 0.5 * (1.0 + std::tanh((x - 1.0) / 0.1));
}
// now a test driver
int main() {
// setup the interpolant and store the points in a file for plotting
int N{11};
double xmin{0.0};
double xmax{2.0};
LagrangePoly l(xmin, xmax, N, f_test);
std::ofstream of("data_samples.txt");
for (std::size_t i = 0; i < l.xp.size(); ++i) {
of << std::setw(14) << l.xp[i] << " " << std::setw(14) << l.fp[i] << std::endl;
}
// now finely sample the domain and interpolate our samples and
// output the interpolation to another file
std::ofstream of2("interpolation.txt");
int N_fine{200};
double dx_fine = (xmax - xmin) / static_cast<double>(N_fine-1);
for (int i = 0; i < N_fine; ++i) {
double x = xmin + static_cast<double>(i) * dx_fine;
auto f_interp = l.evalf(x);
auto f_true = f_test(x);
of2 << std::setw(14) << x << " "
<< std::setw(14) << f_interp << " "
<< std::setw(14) << f_true << std::endl;
}
}


