SciPy ODE Integration#
The python SciPy library includes a lot of different types of ODE integrators, all available through the solve_ivp() interface.
To work on our system, we need to provide a righthand side function of the form:
rhs(t, y)
where y is the vector of unknowns. We’ll assume it is ordered as
\((x, y, u, v)\)
import numpy as np
import matplotlib.pyplot as plt
from scipy import integrate
GM = 4.0 * np.pi**2
def rhs(t, yvec):
x, y, u, v = yvec
dxdt = u
dydt = v
r = np.sqrt(x * x + y * y)
dudt = -GM * x / r**3
dvdt = -GM * y / r**3
return np.array([dxdt, dydt, dudt, dvdt])
Let’s setup the initial conditions
def initial_conditions(a, e):
x0 = 0
y0 = a * (1 - e)
u0 = -np.sqrt(GM / a * (1 + e)/(1 - e))
v0 = 0
return np.array([x0, y0, u0, v0])
Now we can integrate. We have a choice of solvers:
https://docs.scipy.org/doc/scipy/reference/generated/scipy.integrate.solve_ivp.html
We’ll pick RK45, which is the default
a = 1.0
e = 0.3
tmax = 10.0
yvec0 = initial_conditions(a, e)
sol = integrate.solve_ivp(rhs, (0, tmax), yvec0, method="RK45")
The data are returned via the sol object. The two bits we care about the most are t and y, the latter which is an (neqs, nsteps) array.
Since all the data for a single variable is together (row-major storage), this is a struct-of-arrays type layout.
t = sol.t
yvec = sol.y
yvec.shape
(4, 99)
fig, ax = plt.subplots()
ax.plot(yvec[0,:], yvec[1,:])
ax.set_aspect("equal")
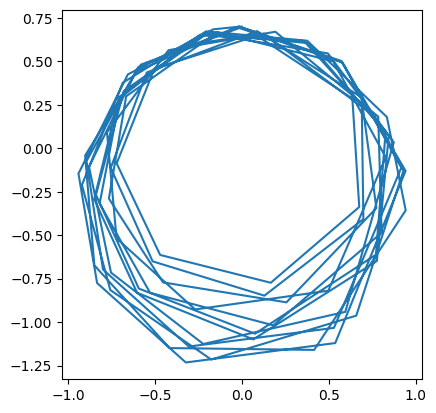
We see that this solution does not look great.
Tip
solve_ivp takes both a relative and absolute tolerance, and their defaults are rtol=1.e-3, atol=1.e-6.
They are combined into an error tolerance of \(\mathtt{atol} + \mathtt{rtol} |y|\).
Tighter tolerances#
Let’s make the tolerances tighter
sol = integrate.solve_ivp(rhs, (0, tmax), yvec0, method="RK45", rtol=1.e-6)
t = sol.t
yvec = sol.y
fig, ax = plt.subplots()
ax.plot(yvec[0,:], yvec[1,:])
ax.set_aspect("equal")
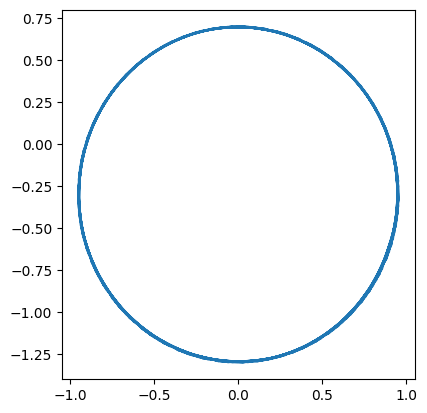
This looks much better.


