OpenMP Poisson Relaxation#
Let’s extend our 2d implementation of the Poisson solver to work with OpenMP. Most of the work is in the smoothing / relaxation routine, so we will tackle that first.
We did Gauss-Seidel relaxation—this means that we use the new values of \(\phi_{i,j}\) as soon as they become available. If we parallelize the loops using OpenMP, then there is a risk that while we are updating a point on one thread another thread will be reading it to use it in its update. This is a race condition. We need to avoid these.
As we saw previously, the 2D update has the form:
If \(\Delta x = \Delta y\), then this simplifies to:
We see that the update of \(\phi_{i,j}\) depends only on the values above, below, and to the left and right. We can visualize this as a checkerboard:
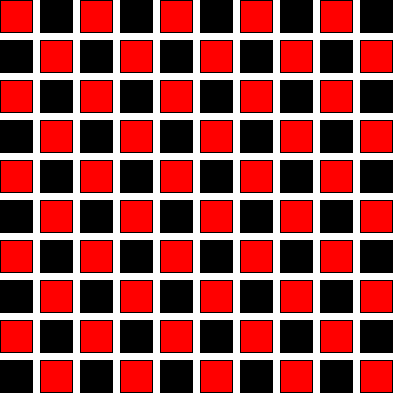
The update of the red points depends only on the black points and vice versa. This means that we can split the update into two separate passes—first updating the red points and then the black. This is called red-black Gauss-Seidel.
Here’s what this code looks like:
double fac = dx * dx * dy * dy / (dx * dx + dy * dy);
double dx2i = 1.0 / (dx * dx);
double dy2i = 1.0 / (dy * dy);
for (int n = 0; n < n_smooth; ++n) {
#pragma omp parallel
{
#pragma omp for
for (int i = 1; i < nx-1; ++i) {
int offset = i % 2;
for (int j = 1 + offset; j < ny-1; j += 2) {
phi(i,j) = 0.5 * fac * (dx2i * (phi(i-1,j) + phi(i+1,j)) +
dy2i * (phi(i,j-1) + phi(i,j+1)) - f(i,j));
}
}
#pragma omp for
for (int i = 1; i < nx-1; ++i) {
int offset = (i + 1) % 2;
for (int j = 1 + offset; j < ny-1; j += 2) {
phi(i,j) = 0.5 * fac * (dx2i * (phi(i-1,j) + phi(i+1,j)) +
dy2i * (phi(i,j-1) + phi(i,j+1)) - f(i,j));
}
}
} // openmp
}
We have a single parallel region and then 2 separate parallel for loops. There is an implicit “wait” after each loop, so we ensure that all threads reach the end before going onto the next loop.
The only other routine that requires a little care is norm, since
we have a reduction there, but we can handle this in the same way we
did previously.
Here’s the full implementation:
poisson2d.H
#ifndef POISSON_H
#define POISSON_H
#include <cassert>
#include <cmath>
#include <limits>
#include <fstream>
#include <iomanip>
#include <functional>
#include <vector>
#include "array.H"
///
/// Solve the 2-d Poisson problem on a node-centered finite-difference
/// grid with Dirichlet BCs using Gauss-Seidel smoothing
///
class Poisson {
private:
double xmin;
double xmax;
double ymin;
double ymax;
int nx;
int ny;
std::vector<double> x;
std::vector<double> y;
Array phi;
Array f;
double dx;
double dy;
public:
Poisson(double xmin_in, double xmax_in,
double ymin_in, double ymax_in,
int nx_in, int ny_in)
: xmin{xmin_in}, xmax{xmax_in}, ymin{ymin_in}, ymax{ymax_in},
nx{nx_in}, ny{ny_in},
x(nx_in, 0), y(ny_in, 0),
phi(nx_in, ny_in, 0), f(nx_in, ny_in, 0)
{
// initialize the coordinates
assert (xmax > xmin);
assert (ymax > ymin);
dx = (xmax - xmin) / static_cast<double>(nx-1);
dy = (ymax - ymin) / static_cast<double>(ny-1);
for (int i = 0; i < nx; ++i) {
x[i] = xmin + static_cast<double>(i) * dx;
}
for (int j = 0; j < ny; ++j) {
y[j] = ymin + static_cast<double>(j) * dy;
}
}
///
/// set the -x Dirichlet boundary condition
///
void set_xlo_bc(double val) {
for (int j = 0; j < ny; ++j) {
phi(0, j) = val;
}
}
///
/// set the +x Dirichlet boundary condition
///
void set_xhi_bc(double val) {
for (int j = 0; j < ny; ++j) {
phi(nx-1, j) = val;
}
}
///
/// set the -y Dirichlet boundary condition
///
void set_ylo_bc(double val) {
for (int i = 0; i < nx; ++i) {
phi(i, 0) = val;
}
}
///
/// set the +y Dirichlet boundary condition
///
void set_yhi_bc(double val) {
for (int i = 0; i < nx; ++i) {
phi(i, ny-1) = val;
}
}
///
/// set the source term, f, in L phi = f
///
void set_source(std::function<double(double, double)> func) {
#pragma omp parallel for collapse(2)
for (int i = 0; i < nx; ++i) {
for (int j = 0; j < ny; ++j) {
f(i,j) = func(x[i], y[j]);
}
}
}
///
/// return the number of points in x
///
int nx_pts() {return nx;}
///
/// return the number of points in y
///
int ny_pts() {return ny;}
///
/// return the x coordinate vector
///
const std::vector<double>& xc() {return x;}
///
/// return the y coordinate vector
///
const std::vector<double>& yc() {return y;}
///
/// return the source vector
///
Array& get_source() {
return f;
}
///
/// return the solution vector
///
Array& get_phi() {
return phi;
}
///
/// do Gauss-Seidel smoothing for n_smooth iterations
///
void smooth(int n_smooth) {
// perform Red-Black Gauss-Seidel smoothing
// we only operate on the interior nodes
double fac = dx * dx * dy * dy / (dx * dx + dy * dy);
double dx2i = 1.0 / (dx * dx);
double dy2i = 1.0 / (dy * dy);
for (int n = 0; n < n_smooth; ++n) {
#pragma omp parallel
{
#pragma omp for
for (int i = 1; i < nx-1; ++i) {
int offset = i % 2;
for (int j = 1 + offset; j < ny-1; j += 2) {
phi(i,j) = 0.5 * fac * (dx2i * (phi(i-1,j) + phi(i+1,j)) +
dy2i * (phi(i,j-1) + phi(i,j+1)) - f(i,j));
}
}
#pragma omp for
for (int i = 1; i < nx-1; ++i) {
int offset = (i + 1) % 2;
for (int j = 1 + offset; j < ny-1; j += 2) {
phi(i,j) = 0.5 * fac * (dx2i * (phi(i-1,j) + phi(i+1,j)) +
dy2i * (phi(i,j-1) + phi(i,j+1)) - f(i,j));
}
}
} // openmp
}
}
///
/// solve the Poisson problem via relaxation until the residual
/// norm is tol compared to the source norm
///
void solve(double tol) {
double err = std::numeric_limits<double>::max();
double source_norm = norm(f);
while (err > tol) {
smooth(10);
double r_norm = norm(residual());
if (source_norm != 0.0) {
err = r_norm / source_norm;
} else {
err = r_norm;
}
}
}
///
/// compute the residual
///
Array residual() {
Array r(nx, ny, 0);
#pragma omp parallel for collapse(2)
for (int i = 1; i < nx-1; ++i) {
for (int j = 1; j < ny-1; ++j) {
r(i,j) = f(i,j)
- (phi(i+1,j) - 2.0 * phi(i,j) + phi(i-1,j)) / (dx * dx)
- (phi(i,j+1) - 2.0 * phi(i,j) + phi(i,j-1)) / (dy * dy);
}
}
return r;
}
///
/// given a vector e on our grid, return the L2 norm
///
double norm(const Array& e) {
double l2{0.0};
#pragma omp parallel reduction(+:l2)
{
#pragma omp for collapse(2)
for (int i = 0; i < nx; ++i) {
for (int j = 0; j < ny; ++j) {
l2 += e(i,j) * e(i,j);
}
}
}
l2 = std::sqrt(dx * dy * l2);
return l2;
}
///
/// give the analytic solution function, return the
/// norm of the true error
///
double true_error(std::function<double(double, double)> true_func) {
Array err(nx, ny);
#pragma omp parallel for collapse(2)
for (int i = 0; i < nx; ++i) {
for (int j = 0; j < ny; ++j) {
err(i, j) = true_func(x[i], y[j]) - phi(i, j);
}
}
return norm(err);
}
///
/// output the solution to file fname
///
void write_solution(const std::string& fname) {
auto of = std::ofstream(fname);
for (int i = 0; i < nx; ++i) {
for (int j = 0; j < ny; ++j) {
of << std::setw(20) << x[i] << " "
<< std::setw(20) << y[j] << " "
<< std::setw(20) << phi(i,j) << std::setw(20) << std::endl;
}
of << std::endl;
}
}
};
#endif
This also uses the array.H class we’ve been developing throughout the semester:
array.H
Here’s a driver for the Poisson solver:
#include <iostream>
#include <chrono>
#include "poisson2d.H"
#include <omp.h>
const double TOL = 1.e-10;
int main() {
std::cout << "total number of threads = " << omp_get_max_threads() << std::endl;
auto start = std::chrono::system_clock::now();
const int N = 256;
auto p = Poisson(0.0, 1.0, 0.0, 1.0, N, N);
p.set_source([] (double x, double y)
{return -2.0 * ((1.0 - 6.0 * x * x) * y * y * (1.0 - y * y) +
(1.0 - 6.0 * y * y) * x * x * (1.0 - x * x));});
p.set_xlo_bc(0.0);
p.set_xhi_bc(0.0);
p.set_ylo_bc(0.0);
p.set_yhi_bc(0.0);
p.solve(TOL);
std::cout << "error = "
<< p.true_error([] (double x, double y)
{return (x*x - x*x*x*x) * (y*y*y*y - y*y);}) << std::endl;
auto end = std::chrono::system_clock::now();
std::cout << "time to solution: " <<
std::chrono::duration_cast<std::chrono::milliseconds>(end -start).count() /
1000.0 << " s" << std::endl;
p.write_solution("poisson.txt");
}
We can compile this as:
g++ -O2 -fopenmp -o poisson poisson.cpp
Tip
Having the compiler optimize here (via -O2) really helps with the
execution.
On my computer (with 8 cores) running with \(128^2\) gives:
|
time (s) |
|---|---|
1 |
1.804 |
2 |
0.939 |
4 |
0.524 |
8 |
0.318 |