Doxygen#
Doxygen is a standard tool for documenting C++ code. It works off of comments added to the code to build documentation (including web pages) describing the structure of a code and its classes and functions.
We’ll continue to work on our cxx-array repo that we just created.
But to help automate things, we need to make a change to how the
webpage is stored.
Using gh-pages#
Our goal is to have GitHub automatically generate our website. When
we do this, it is best to have the actual web page on a separate branch
so we don’t have to keep merging with the regenerating web page
every time we make changes. This is the purpose of the gh-pages
branch. It is an orphaned branch, so it doesn’t have any connection
to main.
Creating the gh-pages branch#
We’ll follow these instructions
for creating the gh-pages branch.
Here’s the procedure. In cxx-array/, make sure you don’t have any
uncommitted work, and then do:
git checkout --orphan gh-pages
git reset --hard
git commit --allow-empty -m "Initializing gh-pages branch"
Now add an index.html in the empty directory:
<!DOCTYPE html>
<html lang="en">
<head>
<title>Hello</title>
<meta charset="utf-8" />
</head>
<body>
<h1>C++ Array Class</h1>
<p><tt>Array.H</tt> provides a simple C++ multi-dimensional array class.</p>
</body>
</html>
Let’s also add the .nojekyll file:
touch .nojekyll
Now let’s commit these:
git add index.html .nojekyll
git commit
Finally, let’s push our branch to our repo:
git push origin gh-pages
Updating the web location on GitHub#
Now we need to go back to the GitHub page for our repo and go to settings and “Pages” and change the “Source” to be “Branch: gh-pages” and “/ (root)” and save.
After a minute or so, the page should be regenerated and appear at the same URL as before.
Note
From now on, when we want to manually make changes to our web page, we need
to be on the gh-pages branch.
We can now switch back to main:
git checkout main
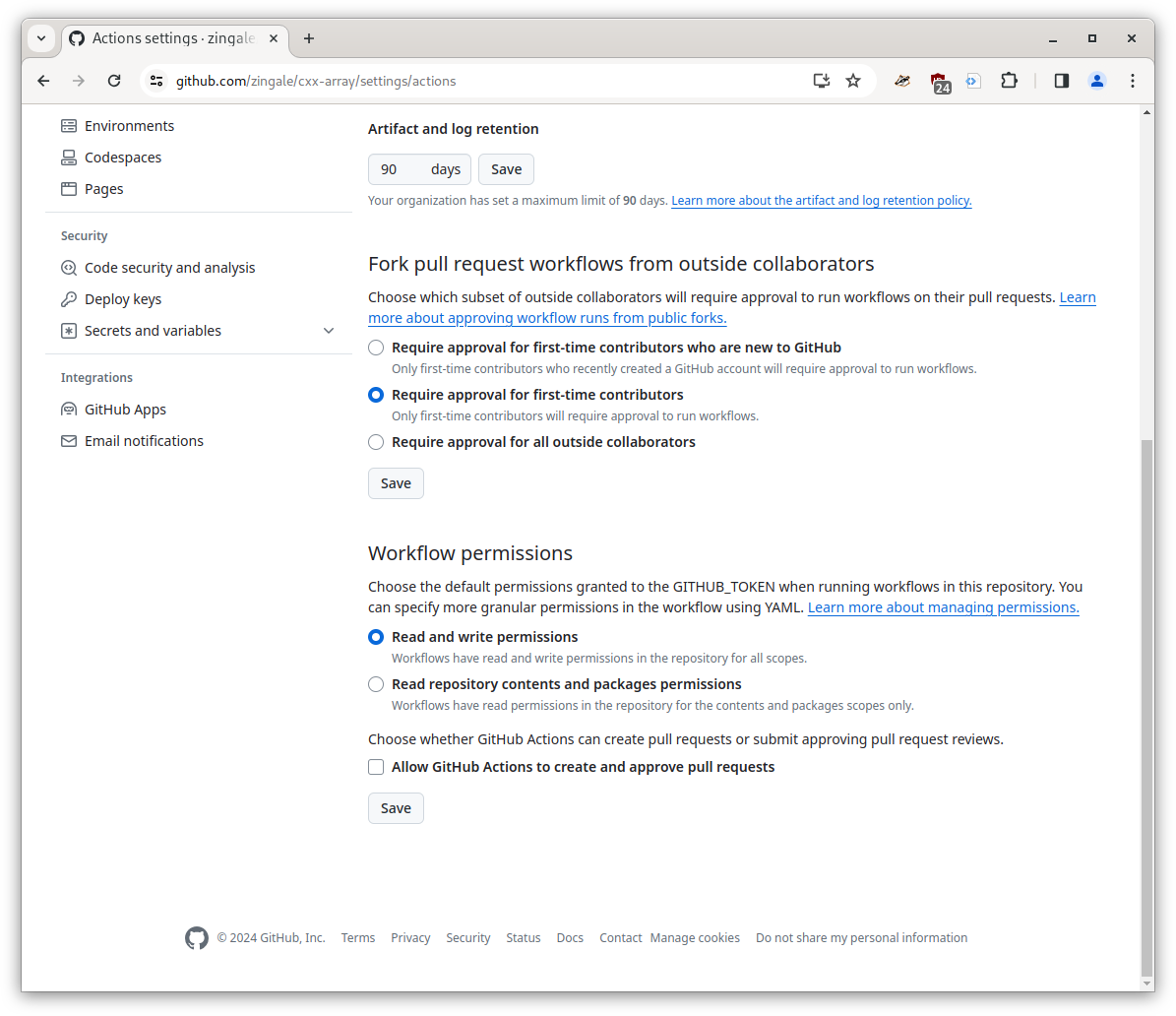
Setting permissions#
We also need to update our github repo to grant an action the ability to write to our repo. Under the settings menu, go to actions settings in the left pane (and pick general). Then navigate to the “workflow permissions” section near the bottom and grant read/write permissions like shown below:
Doxygen configuration#
Setting up: https://www.doxygen.nl/manual/starting.html
doxygen -g
This creates a file called Doxyfile. We need to make a few edits to it:
We want to change
PROJECT_NAMEtoC++ ArrayWe want to change
OUTPUT_DIRECTORYtodocs, since that’s where we expect our source to liveWe’ll have Doxygen use the same
README.mdthat github displays as the main page for the Doxygen start page. This is done by changing:USE_MDFILE_AS_MAINPAGE = README.md
We’ll turn off Latex output by changing
GENERATE_LATEXfromYEStoNOWe’ve been using
*.Hfor our header files, so we need to add this to the listFILE_PATTERNSin theDoxyfile.Doxygen already associates
.Hwith C++, but just doesn’t search for it by default.
Tip
Add Doxyfile to your git repo and commit.
Annotating our code#
Doxygen uses /// as the documentation comment style for C++.
So we would do:
///
/// a function to add two numbers
///
double add(double x, double y);
Note that we have an empty /// before and after the documentation.
Now we can annotate our C++ Array class code. Here’s a version of array.H that we developed in class with
some documentation comments.
#ifndef ARRAY_H
#define ARRAY_H
#include <vector>
#include <iostream>
#include <cassert>
#include <limits>
///
/// a contiguous 2-d array
/// here the data is stored in row-major order in a 1-d memory space
/// managed as a vector. We overload () to allow us to index this as
/// a(irow, icol)
///
class Array {
private:
std::size_t _rows;
std::size_t _cols;
std::vector<double> _data;
public:
Array (std::size_t rows, std::size_t cols, double val=0.0)
: _rows{rows},
_cols{cols},
_data(rows * cols, val)
{
// we do the asserts here after the initialization of _data
// in the initialization list, but if the size is zero, we'll
// abort here anyway.
assert (rows > 0);
assert (cols > 0);
}
// copy constructor
Array (const Array& src) = default;
// assignment operator
Array& operator= (const Array& src) = default;
// destructor
~Array() = default;
// note the "const" after the argument list here -- this means
// that this can be called on a const Array
///
/// get the number of columns
///
inline std::size_t ncols() const { return _cols;}
///
/// get the number of rows
///
inline std::size_t nrows() const { return _rows;}
///
/// get the total number of elements in the array
///
inline std::size_t nelements() const { return _cols * _rows;}
inline double& operator()(int row, int col) {
assert (row >= 0 && row < _rows);
assert (col >= 0 && col < _cols);
return _data[row*_cols + col];
}
inline const double& operator()(int row, int col) const {
assert (row >= 0 && row < _rows);
assert (col >= 0 && col < _cols);
return _data[row*_cols + col];
}
///
/// return a flattened view of the data region -- this can be used with
/// a range-for loop, e.g. `for (auto &e : a.flat()) {...}`
///
inline std::vector<double>& flat() { return _data; }
///
/// set an array element in the flattened index space
///
inline double& flat(int element) {
assert (element >= 0 && element < _rows * _cols);
return _data[element];
}
///
/// get an array element in the flattened index space for
/// a const array
///
inline const double& flat(int element) const {
assert (element >= 0 && element < _rows * _cols);
return _data[element];
}
///
/// return the global minimum of the array
///
double min() const {
double arr_min{std::numeric_limits<double>::max()};
for (const double& e : _data) {
arr_min = std::min(arr_min, e);
}
return arr_min;
}
///
/// return the global maximum of the array
///
double max() const {
double arr_max{std::numeric_limits<double>::lowest()};
for (const double& e : _data) {
arr_max = std::max(arr_max, e);
}
return arr_max;
}
friend std::ostream& operator<< (std::ostream& os, const Array& a);
};
// the << operator is not part of the of the class, so it is not a member
inline
std::ostream& operator<< (std::ostream& os, const Array& a) {
for (std::size_t row=0; row < a.nrows(); ++row) {
for (std::size_t col=0; col < a.ncols(); ++col) {
os << a(row, col) << " ";
}
os << std::endl;
}
return os;
}
#endif
Tip
Add array.H to your git repo and commit.
Trying it out locally#
We can test out Doxygen locally by doing:
doxygen Doxyfile
The HTML will then be in the docs/html/ directory, and we can view the page as:
google-chrome docs/html/index.html
Tip
The default Doxygen generated pages look a little bland. There are themes available to make it look a lot nicer.
Setting up a GitHub action#
Now create a file: .github/workflows/gh-pages.yml with the following content:
name: github pages
on:
push:
branches:
- main
jobs:
deploy:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v4
- name: Install pandoc and doxygen
run: |
sudo apt install pandoc doxygen
- name: Setup python
uses: actions/setup-python@v5
with:
python-version: '3.12'
- name: Install dependencies
run: python3 -m pip install -r ./requirements.txt
- name: Build Doxygen
run: |
mkdir -p docs
doxygen Doxyfile
- name: Deploy
uses: peaceiris/actions-gh-pages@v4
with:
github_token: ${{ secrets.GITHUB_TOKEN }}
publish_dir: ./docs
keep_files: true
This does a little more than we need for now, as it gets us prepped for Sphinx coming next.
Since we’re adding stuff, let’s add a cxx-array/requirements.txt now as well,
with the following content:
sphinx
sphinx_rtd_theme
breathe
Tip
Add requirements.txt and .github/workflow/gh-pages.yml to your git repo and commit.
One last thing…#
We need to go back to gh-pages and now link to the Doxygen docs from our index.html.
We can do that with the following addition to index.html:
<a href="html/index.html">Doxygen documentation</a>
Finally, git push these changes back to GitHub.
voila!
If everything worked correctly, then the GitHub action should run Doxygen and copy the HTML it generated to our web page.