Homework 4 solutions#
import numpy as np
1.#
We start by defining a function to return a Hilbert matrix of size \(N\)
def Hilbert(n):
""" return a Hilbert matrix, H_ij = (i + j - 1)^{-1} """
H = np.zeros((n,n), dtype=np.float64)
for i in range(1, n+1):
for j in range(1, n+1):
H[i-1,j-1] = 1.0/(i + j - 1.0)
return H
Now we’ll loop over the matrix size, construct a Hilbert matrix, initialize our vector \({\bf x}\) as:
and define the righthand side of our linear system as:
and then seek the solution:
for N in range(2, 16):
A = Hilbert(N)
xorig = np.arange(N)
b = A @ xorig
x = np.linalg.solve(A, b)
err = np.max(np.abs(x - xorig))
if N == 2:
print("{:^5} {:^20} {:^20}".format("N", "absolute error", "condition number"))
print("{:5} {:20.10g} {:20.10g}".format(N, err, np.linalg.cond(A, p=1)))
N absolute error condition number
2 0 27
3 2.220446049e-16 748
4 9.874323581e-13 28375
5 3.924416347e-12 943656
6 9.121663425e-10 29070279
7 1.595682519e-08 985194889.6
8 1.505452669e-07 3.387279238e+10
9 2.225784508e-05 1.099651993e+12
10 0.001948411435 3.535684362e+13
11 0.002645431441 1.23453552e+15
12 0.05145843271 4.255909017e+16
13 7.837144291 7.759900159e+17
14 22.9019534 9.835692477e+17
15 18.70912525 1.221413576e+18
We see that when \(N\) is around 13 we have an error that is \(\mathcal{O}(1)\)!
2.#
Let’s start by reading in the data
data = np.loadtxt("signal.txt")
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.plot(data[:,0], data[:,1], label="original data", zorder=100)
ax.plot(data[:,0], data[:,2], label="noisy data")
ax.legend()
<matplotlib.legend.Legend at 0x7ff853f11240>
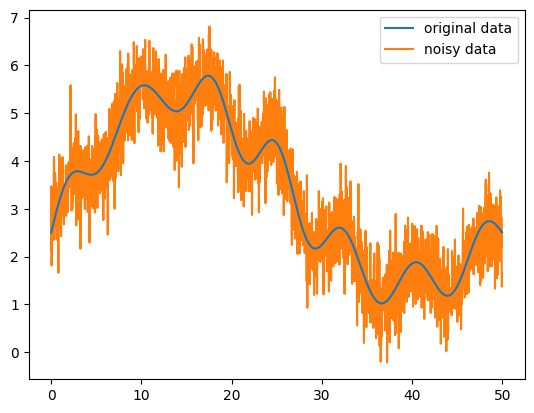
We want to do our best to recover the original data from the noisy signal.
x = data[:, 0]
original = data[:, 1]
signal = data[:, 2]
Let’s define our smoothing function—a Gaussian. We add the function to itself with stride - to get something that is symmetric.
def gaussian(x, sigma=0.5):
""" a gaussian kernel """
g = 1.0/(sigma*np.sqrt(2.0*np.pi))*np.exp(-0.5*(x/sigma)**2)
g = g[:] + g[::-1]
gsum = np.sum(g)
return g/gsum
kernel = gaussian(x)
fig, ax = plt.subplots()
ax.plot(x, kernel)
[<matplotlib.lines.Line2D at 0x7ff853ffb4f0>]
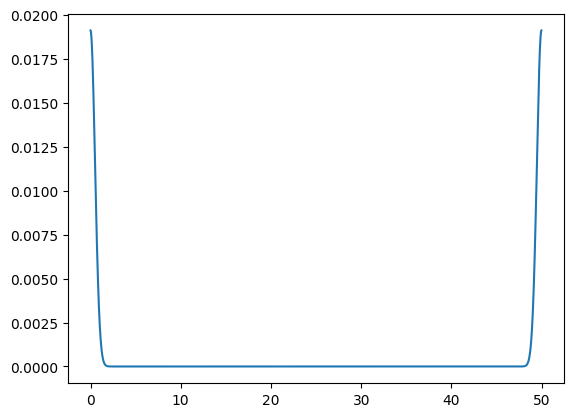
We can see that the amplitude is also quite low—we normalized it such that the discrete data sums to 1 over the domain.
Now let’s take the FFT of both the signal and kernel
fft_signal = np.fft.rfft(signal)
fft_kernel = np.fft.rfft(kernel)
The convolution is just the product of the transforms
conv = fft_signal * fft_kernel
Now let’s transform back
smoothed = np.fft.irfft(conv).real
fig, ax = plt.subplots()
ax.plot(x, original, label="original")
ax.plot(x, smoothed, label="smoothed")
ax.legend()
<matplotlib.legend.Legend at 0x7ff874c4bfa0>
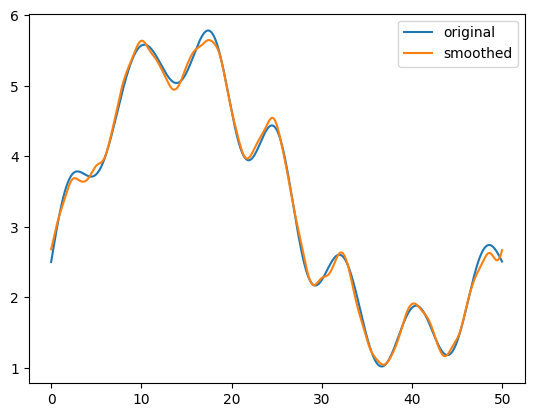
We see that the smoothed data looks quite clean and agrees with the original data very well.


